What It Means to Generate Robust Data in RNA GEP
The goal of most biologic research, including the pursuit of novel therapies to fight disease, is to generate ROBUST data sets so that reliable correlations can be established. But what does “robust” mean when it comes to RNA gene expression profiling (GEP)?
In the world of GEP, robust data begins with in-vitro assays that are resistant to errors when reporting final results. One would expect a robust assay to generate datasets which yield statistically good performance and are largely unaffected by outliers despite the fact that the data come from a wide range of possibility distributions. More specifically, robust datasets demonstrate performance reproducibility, lot-to-lot consistency, high percentage of correlation, and high pass rates.
HTG Transcriptome Panel (HTP) is an example of a robust RNA gene expression profiling assay which possesses the important attributes listed above. HTP was designed to profile the expression of approximately 20,000 mRNA targets and includes over 200 control probes that help ensure consistent and reproducible panel performance.
The Verification Report (P/N C-128.12.R4) of the assay summarizes a Limited Precision study that was performed to assess the precision of the panel across multiple formulation lots, operators, processors, and sample replicates. The assay performance reproducibility and lot-to-lot consistency was measured via Lin’s concordance correlation values which are summarized below.
Such high correlation values across many different sample types confidently demonstrate the robustness of the assay.
Additionally, the White Paper titled “Comparison of Prototype HTG Transcriptome Panel” summarizes how a differential gene expression analysis compares between HTP and traditional RNA-Seq. The results of a Pearson Correlation statistical analysis showed equivalent results using a fraction of the sample input and was able to generate higher sample pass rates when compared to RNA-Seq.
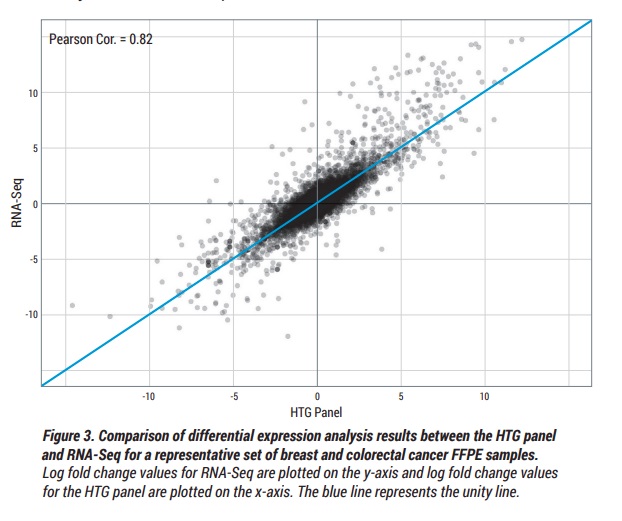
Given the fact that the two assays only share a fraction of the mRNA targets, 82% correlation is considered an indication of a highly robust assay.
Additionally, high pass rates are another indicator of a well-performing and reliable assay. For instance, an assessment of accuracy of the differential gene expression analysis was performed using and HTG Panel and transitional RNA-Seq. After looking at the Pearson correlation coefficients and assessing the log-fold changes of both methods, it was determined that HTG panels exhibit higher pass rates even for archival samples (samples older than 10 years). Below is the summary table of the results of this comparison study.
When selecting an assay for gene expression profiling it is extremely important to choose one that is truly robust. The cost of running an assay with questionable robustness reach far beyond the price of the reagents wasted in the failed run. The cost of the failed run is compounded by specimen scarcity, lost time, project delays, and missed deadlines. Choosing robust and standardized assays is critical to achieving accurate, reliable, and reproducible results in biomedical research.
