HTG EdgeSeq miRNA Whole Transcriptome Assay
The HTG EdgeSeq miRNA Whole Transcriptome Assay (miRNA WTA) enables users to measure the expression of 2,083 human miRNA transcripts using next-generation sequencing (NGS). The HTG EdgeSeq chemistry is compatible with fixed tissue samples, biofluids, and cell lines. The miRNA WTA utilizes our automated HTG EdgeSeq system coupled with the sensitivity and dynamic range of NGS-based detection and can be performed in your lab using the HTG EdgeSeq system, from our VERI/O lab as a service, or at one of the Qualified Service Provider (QSP) sites.
For Research Use Only. Not for Use in Diagnostic Procedures.
Features and Benefits
- Simultaneous, quantitative detection of 2,083 human miRNA transcripts
- Profile circulating miRNA: obtain miRNA profiles from 15 µl of plasma or serum
- Extraction-free: reduce extraction-associated data bias and sample loss
- Gain insights faster: results in as little as 36 hours
- Simplified data analysis: raw numerical data output provided in Excel format
Sample Requirements
| Sample Type | Recommended Sample Input |
|---|---|
Larger sample input amounts must be diluted. Please consult the assay package insert and HTG EdgeSeq System User Manual for more details. |
|
| FFPE Tissue | One 5 µm section |
| Extracted RNA from Frozen | 10 - 35 ng |
| PAXgene | 500 µl |
| Extracted RNA from FFPE | 35 ng |
| Plasma / Serum | ≥15 µl |
| Cells | ≥3,000 cells |
| Extracted RNA | 10 - 35 ng |
Performance
Pearson correlations of >0.95 are routinely obtained between technical replicates of plasma (left chart) and FFPE (right chart).
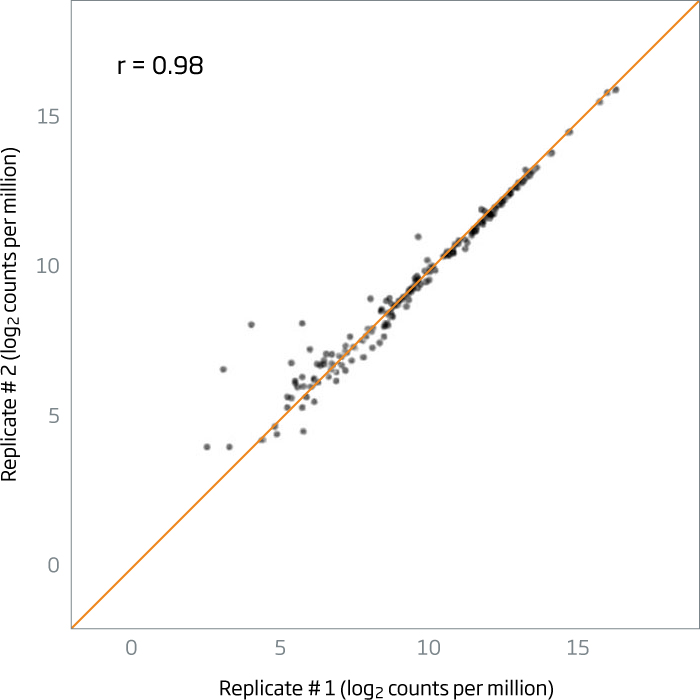
Figure 1
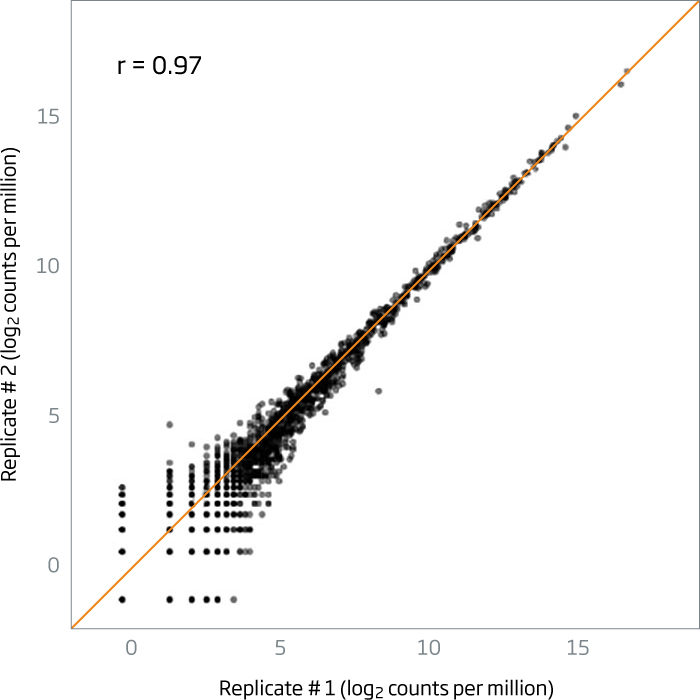
Figure 2
Applications
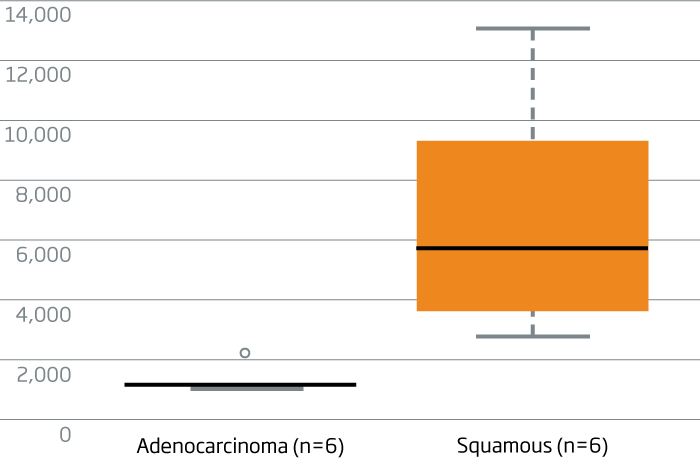
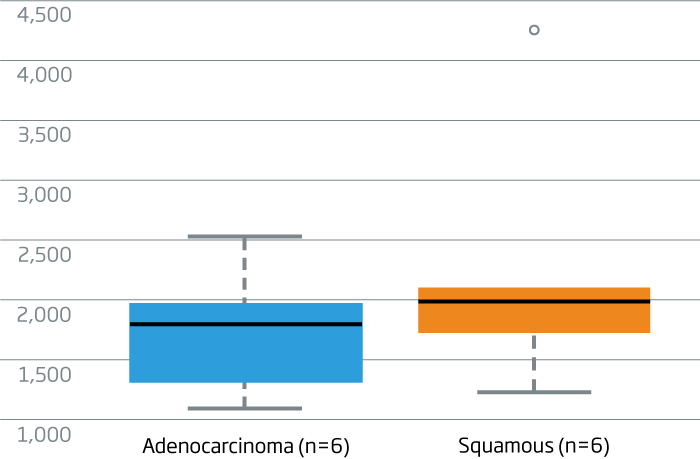
miR-205 is a specific marker for squamous cell carcinoma in lung cancer (Lebanony, et al., 2009). The
HTG EdgeSeq miRNA Whole Transcriptome Assay detects miR-205, differentiating adeno and squamous cell lung carcinomas. A miRNA reported to be expressed in both lung cancer subtypes, let7a-5p, is displayed for reference.
miR-205 expression
let7a-5p expression
Ordering Information
When placing an order, please specify the catalog number. The HTG EdgeSeq miRNA Whole Transcriptome Asssay is compatible with Illumina sequencers and Thermo Fisher Ion Torrent sequencers.
Kit Configurations for use with Illumina Sequencers
916-001-208 HTG EdgeSeq miRNA WTA (2x8)
916-001-008 HTG EdgeSeq miRNA WTA (4x8)
916-001-224 HTG EdgeSeq miRNA WTA (1x24)
916-001-024 HTG EdgeSeq miRNA WTA (4x24)
916-001-096 HTG EdgeSeq miRNA WTA (1x96)
Kit Configurations for use with Thermo Fisher Scientific Ion Torrent Sequencers
916-001-308 HTG EdgeSeq miRNA WTA (2x8)
916-001-108 HTG EdgeSeq miRNA WTA (4x8)
916-001-324 HTG EdgeSeq miRNA WTA (1x24)
916-001-124 HTG EdgeSeq miRNA WTA (4x24)
916-001-196 HTG EdgeSeq miRNA WTA (1X96)
For Research Use Only. Not for Use in Diagnostic Procedures.
Resources and Publications
Learn more about the HTG EdgeSeq miRNA Whole Transcriptome Assay
MicroRNAs in CSF as prodromal biomarkers for Huntington’s disease
View Webinar
Evaluation of a novel microRNA Whole Transcriptome Assay
View Webinar
HTG EdgeSeq miRNA Whole Transcriptome Assay identifies miRNA expression signatures in biofluids aiding in forensic identification
Download pdf 794KB
HTG EdgeSeq miRNA Whole Transcriptome Assay Cross-Species Gene List
Download pdf 238KB
HTG EdgeSeq miRNA Whole Transcriptome Assay Product Sheet
Download pdf 749KB
Publications
TP53 gain-of-function mutation modulates the immunosuppressive microenvironment in non-HPV associated oral squamous cell carcinoma
View External Link
The microRNA Cargo of Human Vaginal Extracellular Vesicles Differentiates Parasitic and Pathobiont Infections from Colonization by Homeostatic Bacteria
View External Link
ASH 2022: 4265: Durable Responses from Acalabrutinib in Combination with Rituximab, Cyclophosphamide, Doxorubicin, Vincristine and Prednisolone (R-CHOP) As First Line Therapy for Patients with Diffuse Large B-Cell Lymphoma (DLBCL): The Accept Phase Ib/II Single Arm Study
View External Link
What Happens In Utero Does Not Stay In Utero: a Review of Evidence for Prenatal Epigenetic Programming by Per- and Polyfluoroalkyl Substances (PFAS) in Infants, Children, and Adolescents
View External Link
Host miRNAs as biomarkers of SARS-CoV-2 infection: a critical review
View External Link
Plasma circulating microRNAs associated with obesity, body fat distribution, and fat mass: the Rotterdam Study
View External Link
Plasma microRNA signature of alcohol consumption: the Rotterdam Study
View External Link
MicroRNA-375 repression of Kruppel-like factor 5 improves angiogenesis in diabetic critical limb ischemia
View External Link
Genome-Wide Profiling of Circulatory microRNAs Associated with Cognition and Dementia
Download pdf 3.0MB
View External Link
A Cross-Comparison of High-Throughput Platforms for Circulating MicroRNA Quantification, Agreement in Risk Classification, and Biomarker Discovery in Non-Small Cell Lung Cancer
Small RNA-Sequencing for Analysis of Circulating miRNAs: Benchmark Study
View External Link
miR-106b-25 Dysregulation in Complex Regional Pain Syndrome Contributes to T Cell Dysfunction
View External Link
Lung Fibrosis Is Improved by Extracellular Vesicles from IFNγ-Primed Mesenchymal Stromal Cells in Murine Systemic Sclerosis
View External Link
Wildfires and extracellular vesicles: Exosomal MicroRNAs as mediators of cross-tissue cardiopulmonary responses to biomass smoke
View External Link
Establishment of a novel multi-omic biomarker panel in cyst fluid and blood for stratifying patient risk of pancreatic cancer
View External Link
Extracellular miR-6723-5p could serve as a biomarker of limbal epithelial stem/progenitor cell population
View External Link
View Resource
T-cell prolymphocytic leukemia is associated with deregulation of oncogenic microRNAs on transcriptional and epigenetic level
View External Link
Placental genomics mediates genetic associations with complex health traits and disease
View External Link
Cellular miR-150-5p may have a crucial role to play in the biology of SARS-CoV-2 infection by regulating nsp10 gene
View External Link
Cell-free plasma microRNAs that identify patients with glioblastoma
View External Link
Blood-based multi-omic signatures of blood-brain barrier impairment and CSF-defined Alzheimer disease in older adults
View External Link
Kikuchi-Fujimoto disease is mediated by an aberrant type I interferon response
View External Link
Increased serum miR-193a-5p during non-alcoholic fatty liver disease progression: diagnostic and mechanistic relevance
View External Link
Multi-omic profiling of patient pancreatic cyst fluid for the identification of a novel biomarker panel of patient cancer risk
View External Link
Circulating MicroRNAs in Relation to Esophageal Adenocarcinoma Diagnosis and Survival
View External Link
Circulating microRNA Breast Cancer Biomarker Detection in Patient Sera with Surface Plasmon Resonance Imaging Biosensor
View External Link
Biomarkers and Lung Cancer Early Detection: State of the Art
View External Link
Comparing the Predictivity of Human Placental gene, microRNA, and CpG Methylation Signatures in Relation to Perinatal Outcomes
View External Link
Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors
Download pdf 4.4MB
View External Link
Kinetics Analysis of Circulating microRNAs Unveils Markers of Failed Myocardial Reperfusion.
Download pdf 3.0MB
Epigenetic-sensitive Pathways in Personalized Therapy of Major Cardiovascular Diseases
View External Link
MicroRNA-based risk scoring system to identify early-stage oral squamous cell carcinoma patients at high-risk for cancer-specific mortality
Download pdf 1.7MB
Synovial-Fluid miRNA signature for Diagnosis of Juvenile Idiopathic Arthritis
Download pdf 1.6MB
Circulating microRNAs in Fabry Disease
Download pdf 1.4MB
Circulating miRNA Profiling of Women at High Risk for Ovarian Cancer
Download pdf 2.2MB
Hepatocyte-Derived Exosomes Promote Liver Immune Tolerance: Possible Implications for Idiosyncratic Drug-Induced Liver Injury
Download pdf 487KB
MicroRNAs: Key Regulators to Understand Osteoclast Differentiation?
Download pdf 4.0MB
Detection of a MicroRNA molecular signature of ultraviolet radiation in the superficial regions of melanocytic nevi on sun-exposed skin
Download pdf 18.9MB
View External Link
MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study
Download pdf 547KB
Developmental profiling of microRNAs in the human embryonic inner ear.
Download pdf 33.3MB
View External Link
Creation and Early Validation of Prognostic miRNA Signatures for Squamous Cell Lung Carcinoma by the SPECS Lung Consortium
View External Link
Circulating miRNAS profile and risk of end-stage renal disease in type 1 diabetes
Download pdf 335KB
HTG EdgeSeq miRNA Whole Transcriptome Assay Platform Performance Metrics Utilizing the MicroRNA (miRQC) Study Specimens
Download pdf 1.1MB
Next-Generation Sequencing (NGS) of Two Independent Cohorts Identifies Eleven Circulating miRNAs for Diagnosis of Nash and Fibrosis
Download pdf 934KB
Validation of our HTG EdgeSeq Platform for accurate and sensitive biomarker discovery
Download pdf 3.2MB
Next-Generation Sequencing (NGS) of Two Independent Cohorts Identifies Eleven Circulating mirNAs for Diagnosis of Nash and Fibrosis
Download pdf 934KB
Verification of miRNA Expression using Nuclease Protection and Targeted Next-Generation Sequencing (NGS)
Download pdf 2.1MB
miRNAs differentially expressed by next-generation sequencing in cord blood buffy coat samples of boys and girls
View External Link
Association of tumor and plasma microRNA expression with tumor monosomy-3 in patients with uveal melanoma
View External Link
Learn More
For further information on the HTG EdgeSeq miRNA Whole Transcriptome Assay, please fill out the registration form below or call us at (877) 507-3259.
Page last updated June 29, 2022