COVID-19 Pathogenesis, Lessons Learned from Lymph Nodes
Date: May 25, 2022
Long-lasting clinical presentation of COVID-19, also called long COVID, is characterized by a wide range of symptoms that appear in a significant number of COVID-19 patients after recovery. Understanding the pathogenesis of COVID-19 is therefore critical to the development of effective therapeutics and predicting immune response in patients.
Autopsy studies of COVID-19 casualties can shed light on the disease, it’s effect on human organism, and potential treatments. Dr. Alexandar Tzankov and his team at the University of Medical Genetics at University Hospital Basel, Switzerland conducted a comprehensive study of autopsy samples collected from March to April 2020, to better understand the pathophysiology and gene expression regulation.
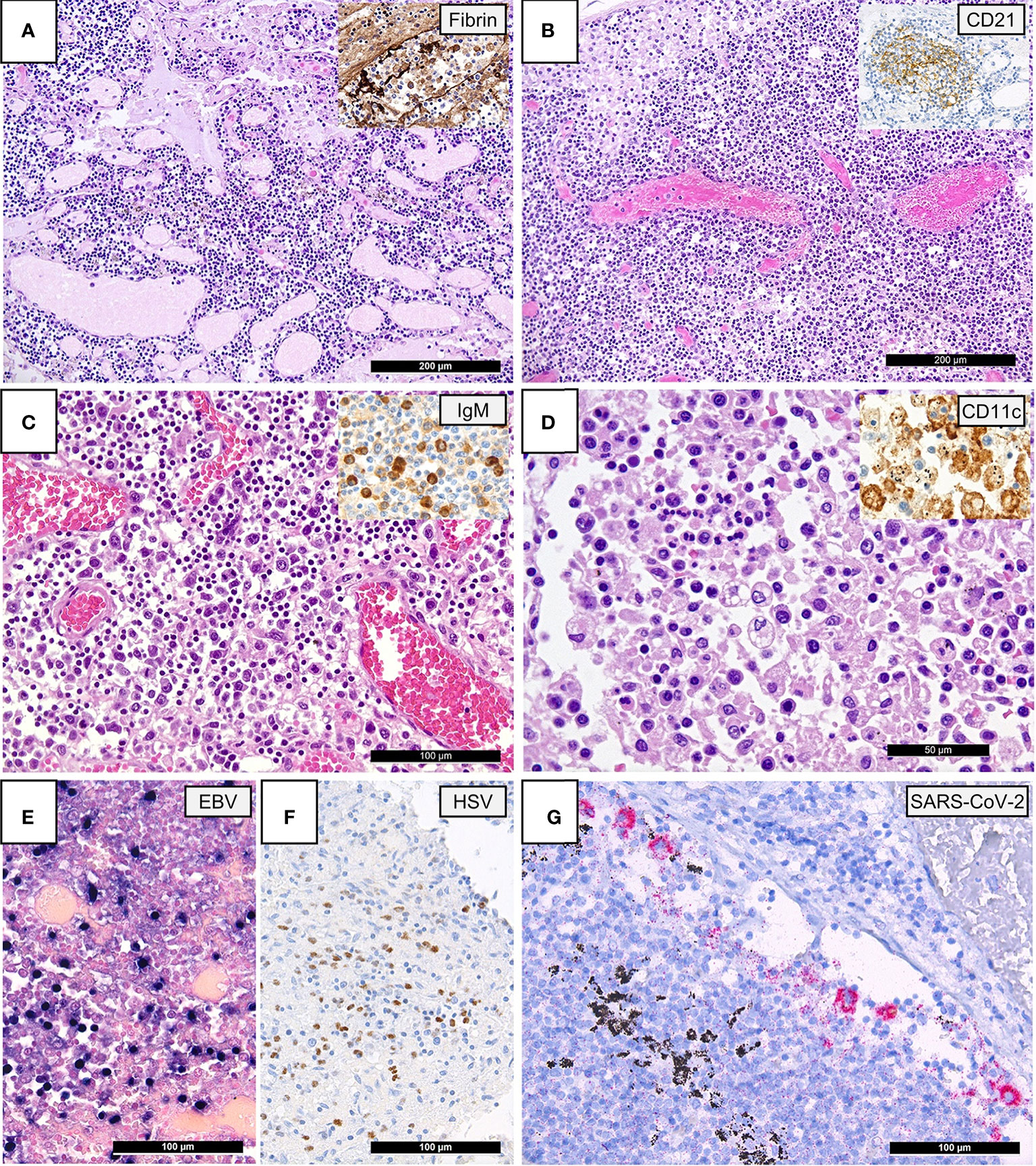
Figure 1: Lymph Node Histomorphology in COVID-19.
(A) Overview of a lymph node draining a COVID-19 lung with edema and capillary stasis (H&E; 100x); inset: fibrin microthrombus in a dilated subcapsular sinus (immunoperoxidase; 100x). (B) Severe capillary stasis and expansion of the paracortex without discernable germinal centers (H&E; 100x); inset: disrupted, CD21+ germinal center network (immunoperoxidase; 100x). (C) Proliferation of extrafollicular plasmablasts (H&E; 200x); inset: expression of IgM by plasmablasts (immunoperoxidase; 360x). (D) Hemophagocytosis in the sinus of a lymph node (H&E; 360x); inset: positivity for CD11c in histiocytes with hemophagocytosis (immunoperoxidase; 400x). (E) Increased amount of EBV-infected B-cells in a lymph node of a COVID-19 patient (EBER ISH; 280x). (F) Increased amount of HSV infected B-cells (immunoperoxidase; 280x). (G) SARS-CoV-2 positivity in sinus histiocytes as detected by immunohistochemistry for the SARS-CoV-2 N-antigen (immunoperoxidase with 3-amino-9-ethylcarbazole used as a chromogen; 280x).
With use of HTG EdgeSeqTM technology the researchers were able to uncover unexpected gene expression profiles such as overexpression of genes related to platelet/vascular functions and angiogenesis and downregulation of genes related to T-/NK functions, leukocyte adhesion and migration. These findings can lead to development of more targeted treatment options in the future.
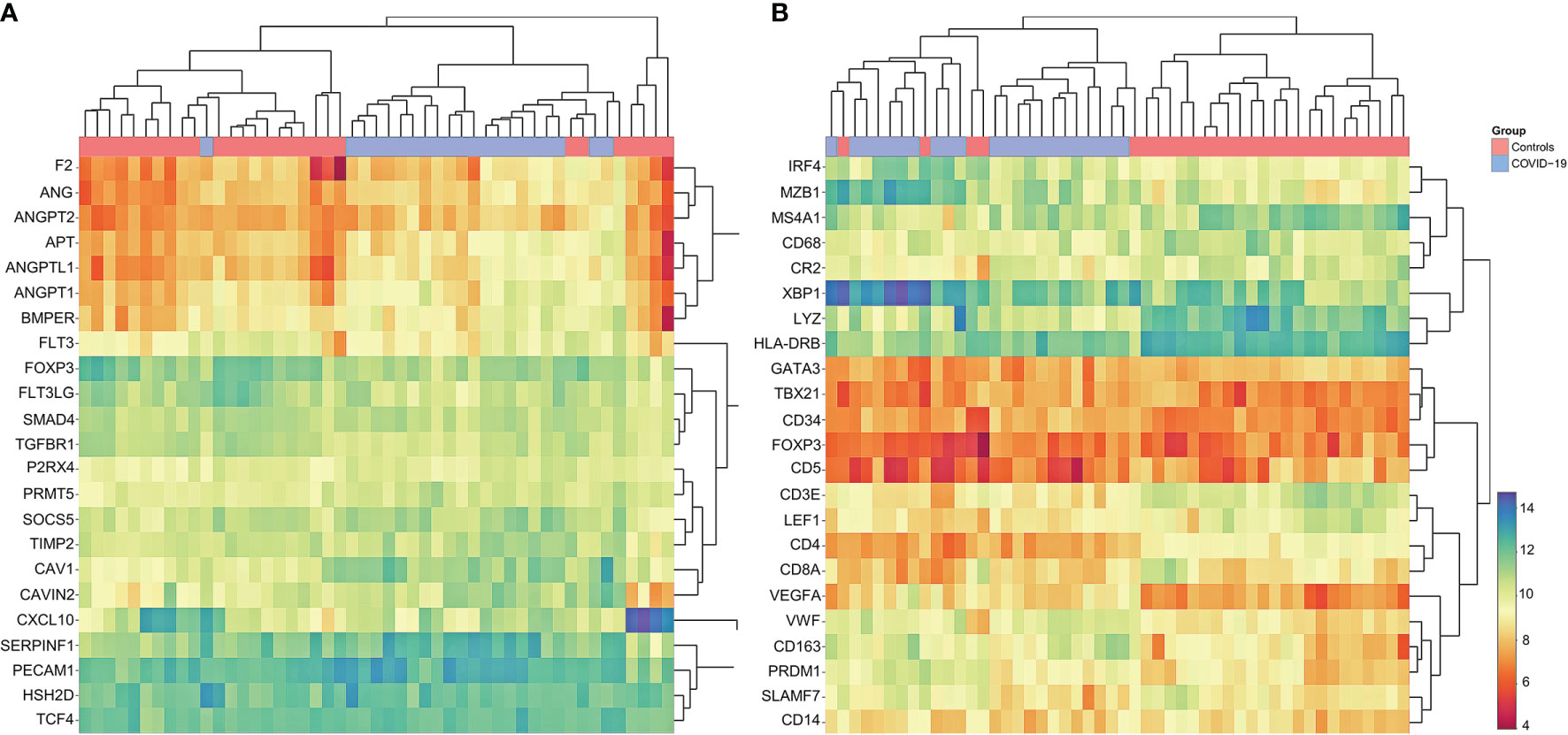
Figure 6: Cluster Analysis of Gene Expression Profiling in COVID-19
(A) Marked dysregulation of angiogenesis-related genes in COVID-19 compared to controls; upregulation of ANG, ANGPT2, AGT, ANGPTL1 ANGPT1, BMPER, SERPINF1 and PECAM1; note that COVID-19 cases cluster together, indicating concordance between individual cases. Individual p-values to be found in Supplementary Materials 2.5. (B) Customized gene set enrichment and clustering analysis of related markers analyzed also by immunohistochemistry shows distinct clustering between COVID-19 and controls: downregulation of CR2 (encodes CD21), TBX21 (encodes Tbet) and LEF1; upregulation of plasmablast genes IRF4, MZB1, XBP1, PRDM1 and SLAMF7; upregulation of angiogenesis related genes (CD34 and VEGF) and of VWF; downregulation of LYZ (encoding for lysozyme) and upregulation of CD163 indicating M2 polarization of macrophages. Note that COVID-19 cases cluster together, indicating concordance between individual cases.
HTG EdgeSeq Immune Response Panel (IRP)
The HTG EdgeSeq IRP used in the publication measures 2,002 immune response mRNA targets in a single RNA extraction-free panel. The list of genes that the panel includes can be found here. The panel leverages the sensitivity and dynamic range of next-generation sequencing (NGS) to measure genes implicated in immune response including response to pathogens such as the SARS-CoV-2 virus as well as dysregulation resulting from autoimmune diseases. The HTG EdgeSeq IRP enables multiplex profiling from FFPE and PAXgene samples.
Webinar
Professor Tzankov, in collaboration with HTG, participated in a webinar where he explained the findings of his team in the abovementioned article. In this video recording of the webinar you will hear about his first-hand experience including:
- gene expression profiling of challenging FFPE tissue samples collected from patient autopsies
- overexpression of genes related to platelet/vascular functions and angiogenesis & downregulation of genes related to T-/NK functions, leukocyte adhesion and migration
- using GEP to develop more targeted treatment options for patients
Resources
The full article and the supplimental materials can be accessed by clicking here.
