Technology
Rethink Possible with the HTG EdgeSeq Workflow and Chemistry
Our technology has a number of key differentiators, including multiplexing from hundreds to thousands of biomarkers in a variety of samples, including formalin-fixed, paraffin-embedded (FFPE) tissue; very low sample input requirements; simple, extraction-free sample preparation; automated workflow with sample-to-result turnaround times in 2 days; and simplified data output. In addition, our HTG EdgeSeq technology generates a molecular profiling library for detection using next‑generation sequencing (NGS). NGS provides improved sensitivity and dynamic range for our HTG EdgeSeq assays.
HTG EdgeSeq Chemistry
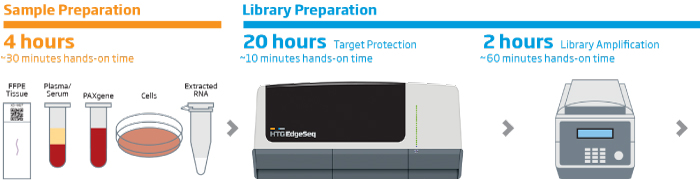
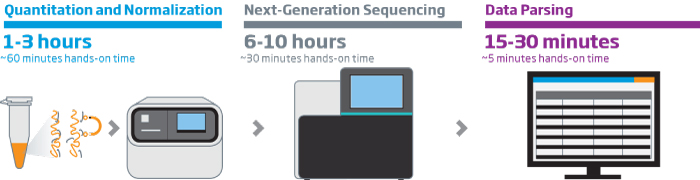
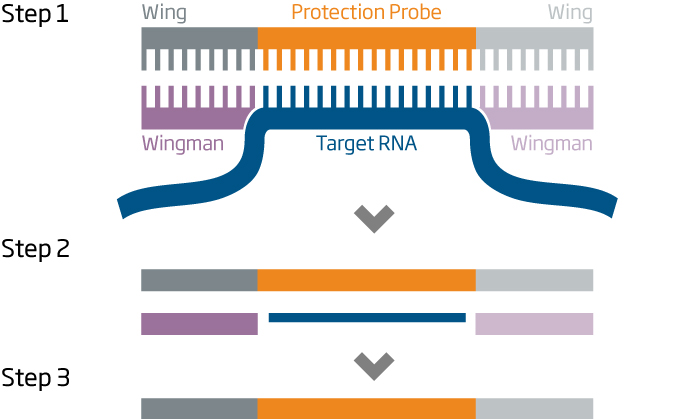
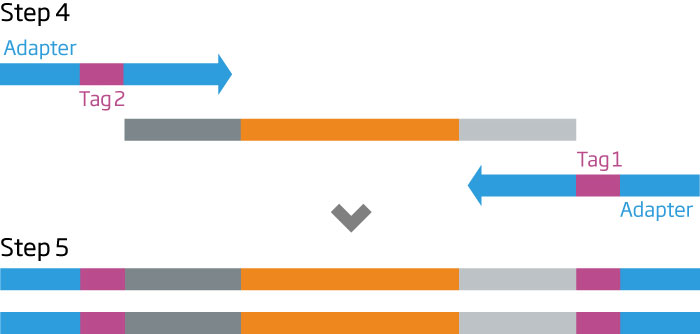
The HTG EdgeSeq chemistry delivers a simple, sample and library preparation method for sequencing-based quantitative assays. The schematic below provides an overview of the chemistry steps from sample to library prep with addition of tags (molecular barcodes) and sequencing adapters. The pooled libraries may be sequenced using most NGS platforms.
HTG EdgeSeq Workflow
Our solutions enable fast, reliable, reproducible, and quantitative analysis of hundreds or thousands of targeted RNAs from a single assay in as little as 36 hours with reduced hands-on time. HTG ‘s extraction-free chemistry significantly reduces sample input requirements and preserves valuable specimens. The HTG EdgeSeq system simplifies NGS library preparation workflow and eliminates time consuming and technically challenging steps like cDNA synthesis, size selection, removal of rRNA, mRNA end repair, and adapter ligation resulting in more reproducible results. HTG’s intuitive graphic user interface simplifies system setup and data analysis.
HTG + Medical Center Core Labs
Now you can use HTG's next-generation sequencing based technology for molecular profiling. Core Labs will process your samples on-site - including sample prep, sequencing, and data analysis, including two-group differential expression analysis. HTG EdgeSeq technology allows you to profile hundreds to thousands of targeted RNAs and miRNAs with no extraction - from most samples as small as:
- 15 μl of biofluid
- One 5 μm FFPE section
- 10 ng of extracted RNA
- ≥3,000 cells
Now available in the following locations:
- Rockville, MD Core Lab + HTG
- Houston, TX - MD Anderson Cancer Center - Sequencing & ncRNA program + HTG
- Chapel Hill, NC - Biomedical Core Facilities, UNC + HTG
- Santa Monica, CA - John Wayne Cancer Institute Sequencing Center + HTG
- Falls Church, VA - Inova Genomics Lab, Inova Translational Medical Institute + HTG
- Denver, CO - University of Colorado Genomics Shared Resource + HTG
To learn more, call our VP, Scientific Collaborations, BJ Kerns at 608-658-0403 or email bjkerns@htgmolecular.com
HTG EdgeSeq Assays
HTG offers a variety of profiling assays for both research and diagnostic uses.
Research Use Only (RUO) Assays
Page last updated November 22, 2019
Ready to get started?
HTG is focused on delivering exceptional molecular profiling products and services. Contact us to find out how our unique technology can benefit you.